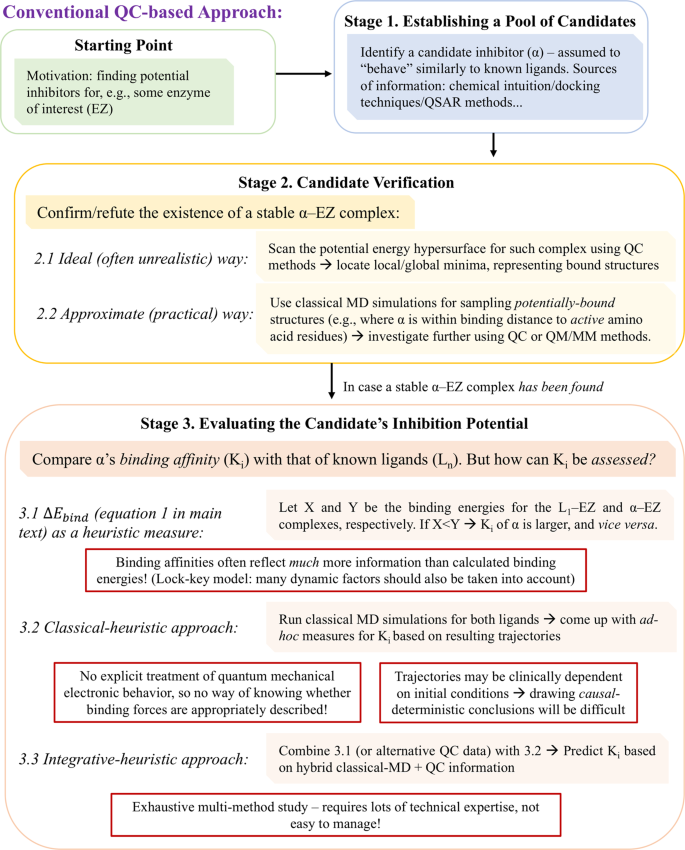
Toward Simple, Predictive Understanding of Protein-Ligand Interactions: Electronic Structure Calculations on Torpedo Californica Acetylcholinesterase Join Forces with the Chemist's Intuition | Scientific Reports
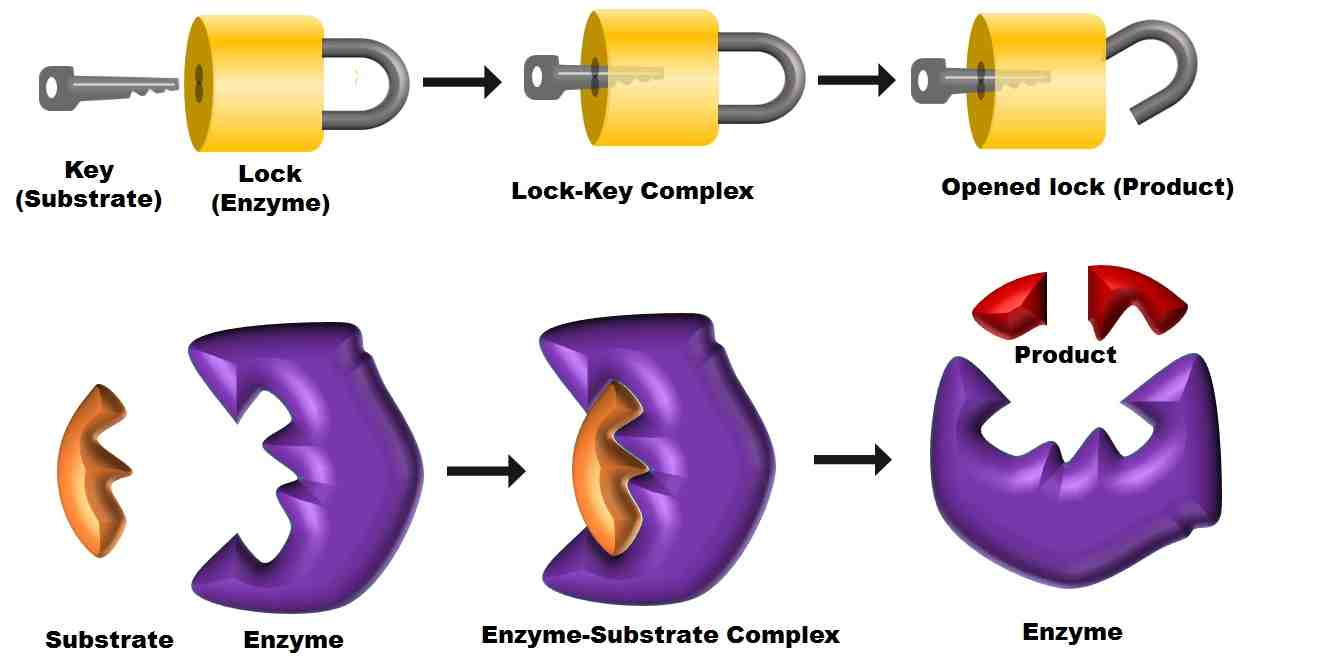
Mechanism of Enzyme Action (Activation Energy and Lock and Key Hypothesis Diagram) - Biology Brain Mechanism of enzyme action
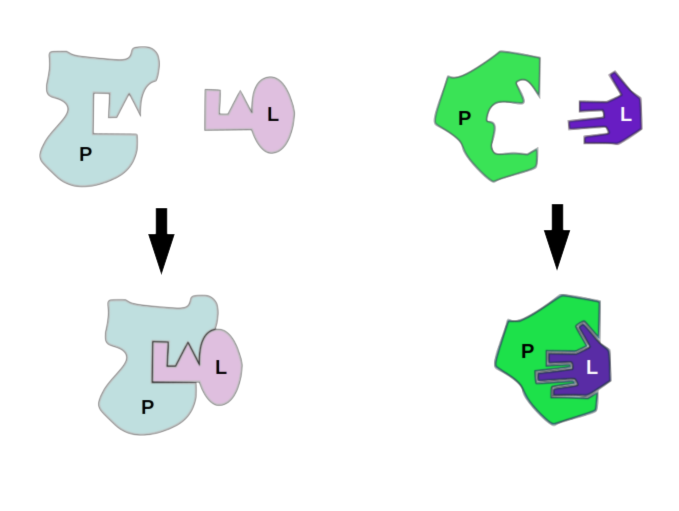
Docking Proteins to Deny Disease: Computational Considerations for Simulating Protein-Ligand Interaction | Exxact Blog

Different technique for Molecular docking: a) Induced work docking b)... | Download Scientific Diagram

Molecular Docking as a Tool to Examine Organic Cation Sorption to Organic Matter | Environmental Science & Technology
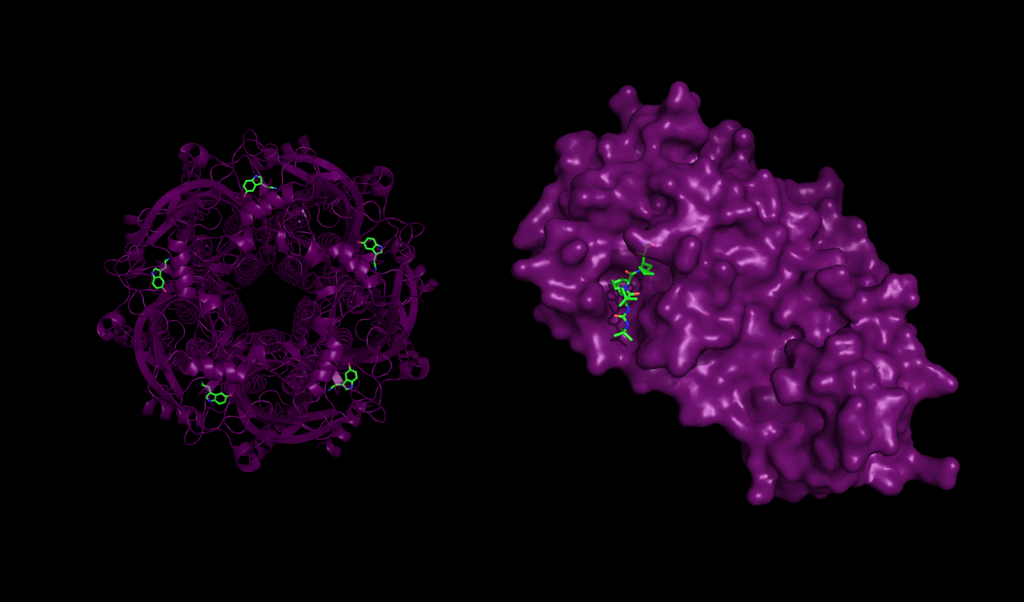
Docking Proteins to Deny Disease: Computational Considerations for Simulating Protein-Ligand Interaction | Exxact Blog

The In Silico Fischer Lock-and-Key Model: The Combined Use of Molecular Descriptors and Docking Poses for the Repurposing of Old Drugs | SpringerLink
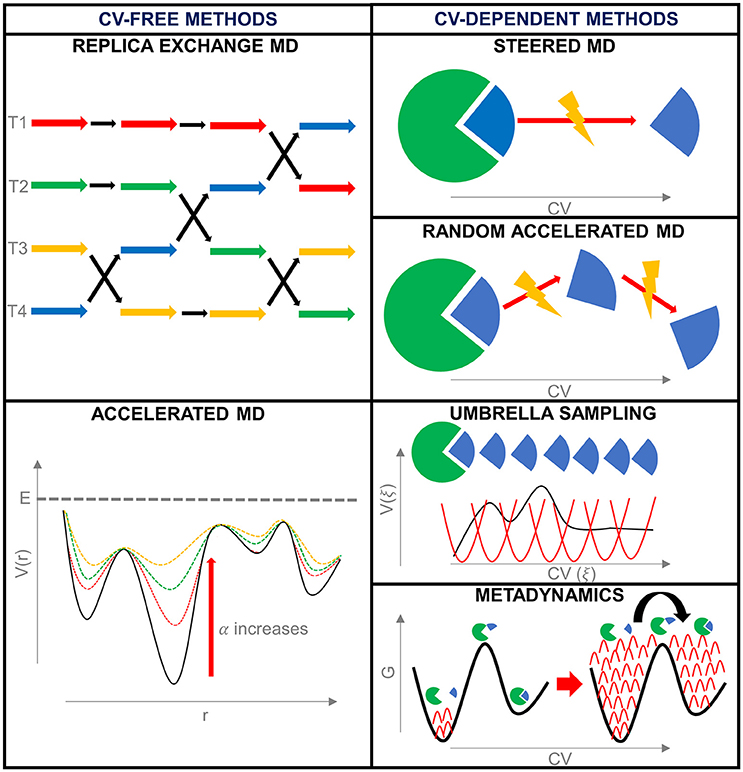
Frontiers | Bridging Molecular Docking to Molecular Dynamics in Exploring Ligand-Protein Recognition Process: An Overview

Research Techniques Made Simple: Molecular Docking in Dermatology - A Foray into In Silico Drug Discovery - ScienceDirect

Research Techniques Made Simple: Molecular Docking in Dermatology - A Foray into In Silico Drug Discovery - ScienceDirect

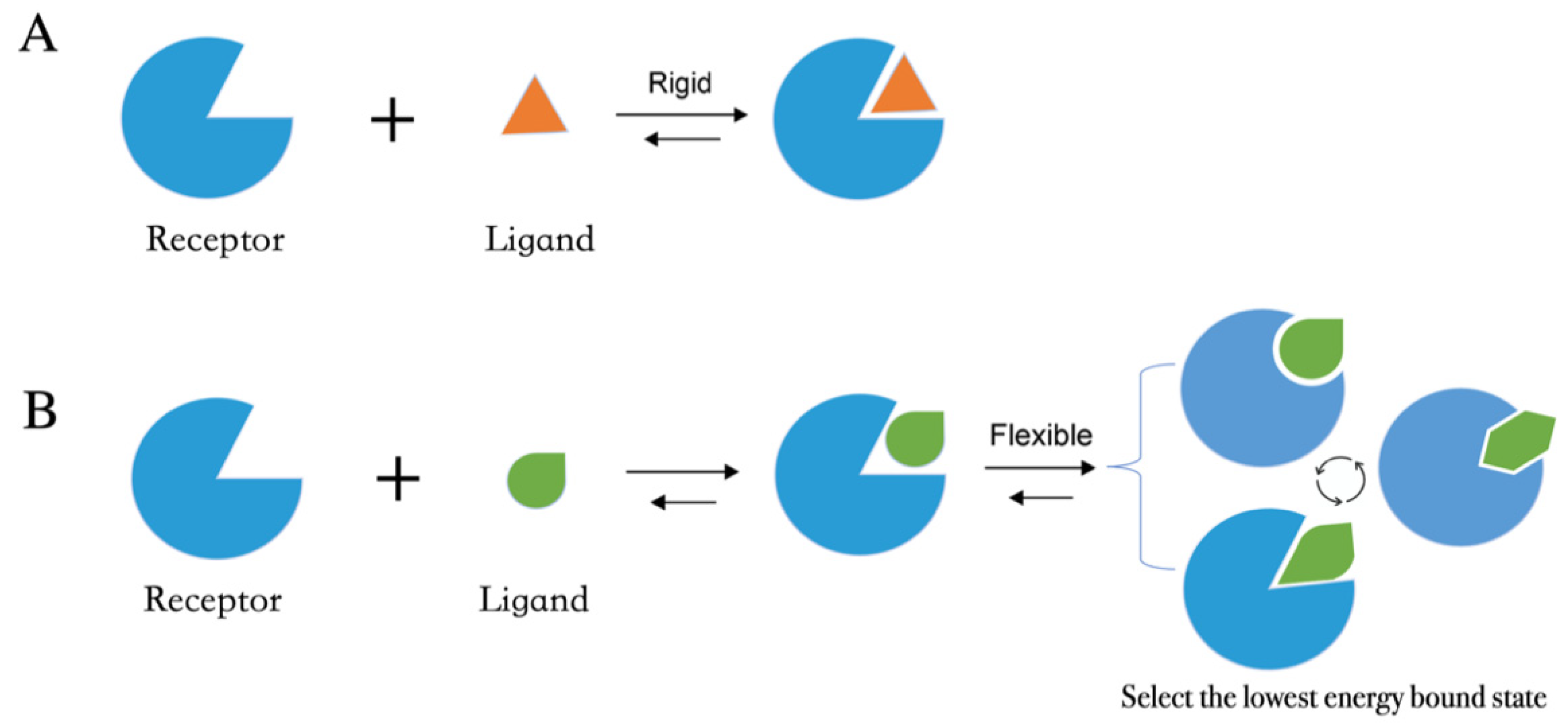
Receptor flexibility in small‐molecule docking calculations - Kokh - 2011 - WIREs Computational Molecular Science - Wiley Online Library




![PDF] Molecular Docking: From Lock and Key to Combination Lock. | Semantic Scholar PDF] Molecular Docking: From Lock and Key to Combination Lock. | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/440d47cf86b98dc01689c8fb3b49815334782c3e/2-Figure2-1.png)